PlotSpec: a tool for generating Gaussian broadened spectra from $tddft calculations
When the user calculates the absorption or emission spectrum of a species using TDDFT, what the user obtains from the BDF program is a list of excitation energies and oscillator strengths in the .out file, for example (C60 molecule, PBE0/def2-SVP, first 10 T1u states):
No. Pair ExSym ExEnergies Wavelengths f D<S^2> Dominant Excitations IPA Ova
1 T1u 1 T1u 3.6161 eV 342.87 nm 0.0539 0.0000 79.2% CV(0): Hu( 26 )-> T1g( 7 ) 4.228 0.724
2 T1u 2 T1u 4.0346 eV 307.30 nm 0.5798 0.0000 58.7% CV(0): Hg( 46 )-> T1u( 19 ) 4.396 0.707
3 T1u 3 T1u 5.2174 eV 237.63 nm 2.4506 0.0000 63.5% CV(0): Hu( 26 )-> Hg( 51 ) 5.475 0.693
4 T1u 4 T1u 5.8548 eV 211.77 nm 0.0055 0.0000 85.7% CV(0): Hg( 41 )-> T1u( 19 ) 6.866 0.566
5 T1u 5 T1u 5.9297 eV 209.09 nm 0.0025 0.0000 72.1% CV(0): Hg( 46 )-> T2u( 19 ) 6.749 0.746
6 T1u 6 T1u 6.2930 eV 197.02 nm 3.3308 0.0000 62.5% CV(0): Fg( 21 )-> T2u( 19 ) 6.724 0.706
7 T1u 7 T1u 6.9435 eV 178.56 nm 0.0027 0.0000 85.6% CV(0): Hu( 21 )-> T1g( 7 ) 7.789 0.539
8 T1u 8 T1u 7.0070 eV 176.94 nm 1.0193 0.0000 87.4% CV(0): Hu( 26 )-> Fg( 25 ) 7.751 0.662
9 T1u 9 T1u 7.5243 eV 164.78 nm 0.0102 0.0000 91.5% CV(0): Hg( 36 )-> T1u( 19 ) 8.351 0.645
10 T1u 10 T1u 7.7409 eV 160.17 nm 0.0145 0.0000 75.0% CV(0): Hg( 46 )-> Hu( 31 ) 8.596 0.599However, what the user really wants is frequently a continuous spectrum that properly reflects the non-zero widths of the peaks, similar to what is typically observed in experiments; the continuous spectrum is then usually plotted together with a set of vertical bars denoting the excitation energies and oscillator strengths of the excited states. This is what the tool plotspec.py (located under $BDFHOME/sbin/) is for. Given a .out file from a TDDFT job (e.g. C60.out), run
$BDFHOME/sbin/plotspec.py C60.out
or more simply
$BDFHOME/sbin/plotspec.py C60
The following information is printed to standard output:
==================================
P L O T S P E C
Spectral broadening tool for BDF
==================================
BDF output file: C60.out
1 TDDFT output block(s) found
Block 1: 10 excited state(s)
- Singlet absorption spectrum, spin-allowed
The spectra will be Gaussian-broadened (FWHM = 0.5000 eV) ...
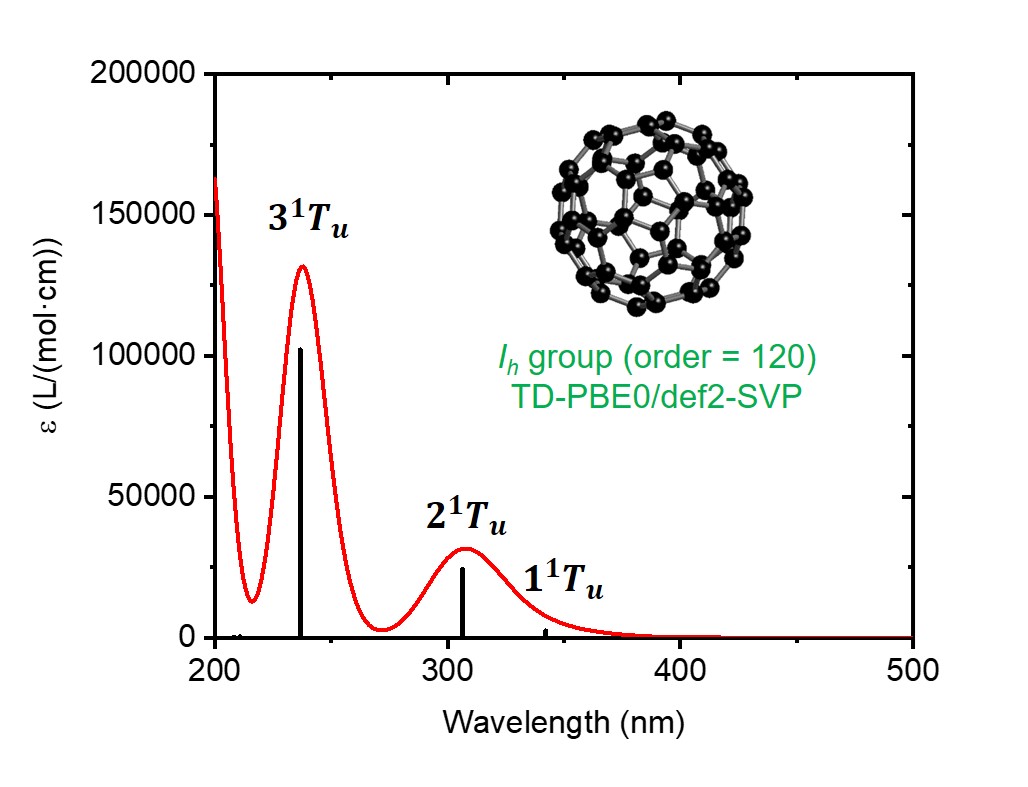
Absorption maxima of spectrum 1 (nm (lg epsilon/(L/(mol cm)))):
- 238 (5.12), 308 (4.50)
plotspec.py: exit successfullyThe script generates two files. One of them is C60.stick.csv, which contains a list of wavelengths and extinction coefficients:
TDDFT Singlets 1,, Wavelength,Extinction coefficient, nm,L/(mol cm), 342.867139,2899.779319, 307.302300,31192.802393, 237.635960,131840.430395, 211.765024,295.895849, 209.090150,134.498113, 197.019205,179194.526059, 178.561512,145.257962, 176.943322,54837.570677, 164.778366,548.752301, 160.167663,780.089056,
The other is C60.spec.csv, which contains the Gaussian broadened spectrum (default FWHM=0.5eV):
TDDFT Singlets 1,, Wavelength,Extinction coefficient, nm,L/(mol cm), 200.000000,162720.545118, 201.000000,151036.824457, 202.000000,137429.257570, ... 998.000000,0.000000, 999.000000,0.000000, 1000.000000,0.000000,
One can then use the two files to plot the predicted absorption spectrum with e.g. Excel or Origin:
A few command line parameters can be specified to fine-tune the behavior of the script. Examples:
Plot the spectrum in the range 300-600 nm: $BDFHOME/sbin/plotspec.py wavelength=300-600nm filename.out Plot an X-ray absorption spectrum in the range 200-210 eV, using a FWHM of 1 eV: $BDFHOME/sbin/plotspec.py energy=200-210eV fwhm=1eV filename.out Plot a UV-Vis spectrum in the range 10000 cm-1 to 40000 cm-1, where the wavenumber is sampled at an interval of 50 cm-1: $BDFHOME/sbin/plotspec.py wavenumber=10000-40000cm-1 interval=50 filename.out Plot an emission spectrum in the range 600-1200 nm, as would be given by Kasha's rule (i.e. only the first excited state is considered), where the wavelength is sampled at an interval of 5 nm: $BDFHOME/sbin/plotspec.py -emi wavelength=600-1200nm interval=5 filename.out
When the file filename.out contains the output of multiple TDDFT jobs (therefore multiple spectra), the spectra are plotted together, as different columns of the files filename.stick.csv and filename.spec.csv. For example, the .spec.csv file from a SOC-corrected transition metal L-edge X-ray absorption spectrum calculation may look like:
TDA Quartets 1,,TDA 6-tets 2,,TDA/TDDFT Spinors 3,, Energy,Extinction coefficient,Energy,Extinction coefficient,Energy,Extinction coefficient, eV,L/(mol cm),eV,L/(mol cm),eV,L/(mol cm), 576.000000,0.000000,576.000000,0.000000,576.000000,0.002005, 576.010000,0.000000,576.010000,0.000000,576.010000,0.002223, 576.020000,0.000000,576.020000,0.000000,576.020000,0.002464, ... 583.000000,2.611833,583.000000,0.000000,583.000000,122.580597, 583.010000,2.607841,583.010000,0.000000,583.010000,121.057743, 583.020000,2.610622,583.020000,0.000000,583.020000,119.510008, ... 591.980000,0.000000,591.980000,0.000000,591.980000,0.069449, 591.990000,0.000000,591.990000,0.000000,591.990000,0.063557, 592.000000,0.000000,592.000000,0.000000,592.000000,0.058133,
In this case the last column contains the SOC-corrected spectrum; the second column contains the spectrum without SOC correction (i.e. the spectrum from the first TDDFT block of the job).
For more detailed options, run $BDFHOME/sbin/plotspec.py without any command line parameter to show the full usage.