BDF User's guide
Insert introduction of BDF module at here.
How to run BDF
To run BDF, you can write a shell script named "run.sh" with following content,
#!/bin/bash # Set BDF home directory export BDFHOME=~/work/0.5.dev # run BDF driver with input file $1 $BDFHOME/sbin/bdfdrv.py -r $1
For example, you can copy the file named "$BDFHOME/Tests/input/test002.inp" to a work directory. Then, you write done the shell script and store it in you work directory. To evoke BDF calculation, you just use command
$./run.sh test001.inp
The output will be printed on standard output. Thus, it is better to redirect output to a file.
$./run.sh test001.inp > test001.out
Some tips to run BDF
1. There are a lot of testing inputs saved in directory of $BDFHOME/Tests/input.
2. BDF driver assume input file has the name *.inp. Thus, you can run BDF with command
$./run.sh test001
3. If BDF is compiled with OpenMP supporting, you can can set OpenMP environmental variables in running script. For example,
export OMP_NUM_THREADS=4
export OMP_STACKSIZE=1024M
BDF Flowchat
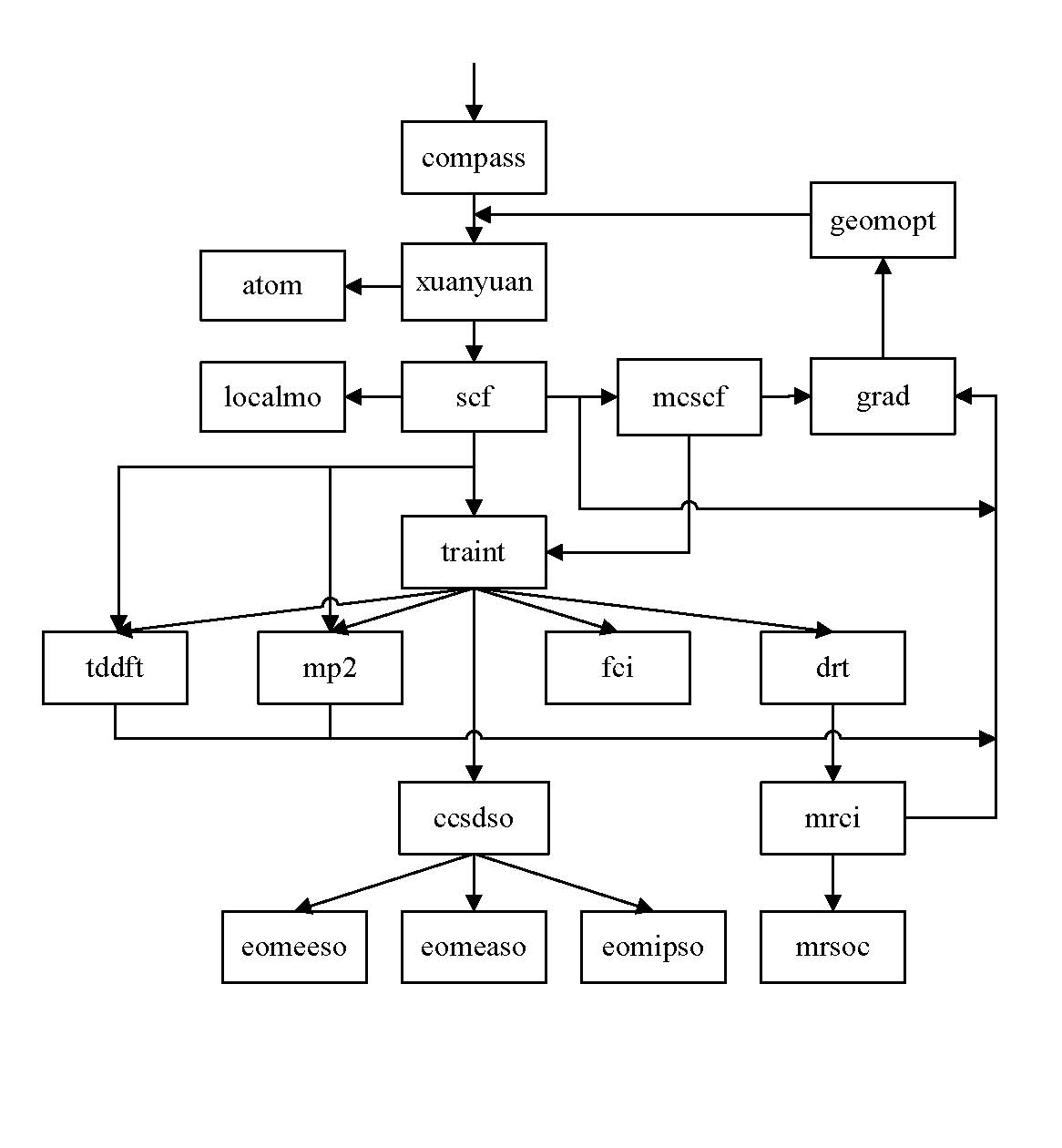
Input style
BDF modules
compass - Molecule geometry and basis set preprocess.
drt - Generate DRTs in GUGA.
grad - Gradient.
mcscf - Multi-configuration self-consistent-field program
mp2 - MP2 program
mrci - Multi-reference configuration interaction program.
localmo - Localization of molecule orbital.
scf - Self-consistent-field program.
tddft - Time dependent density functional program.
vgmfci - electron-nucleus mean field configuration interaction program.
xuanyuan - 1e and 2e integrals program.
genfrag - Generate or optimize fragments and fragments pairs in Local orbital based Frag-MP2/CCSD.
expandmo - Expand molecular orbital from small basis set to large basis set.